New study reveals FTLD-TDP subtypes have distinct epigenetic signatures
The Rademakers lab presents the most comprehensive analysis of DNA methylation patterns in FTLD-TDP to date, revealing that the molecular differences between disease subtypes run even deeper than previously recognized.

Frontotemporal dementia is the second most common form of dementia in people under 65, encompassing a group of diseases that affect the brain's frontal and temporal lobes, causing changes in speech, behavior, and personality. While scientists have identified three main disease subtypes (Type A, B, and C) based on where TDP-43 protein clumps appear in brain tissue and what they look like, the majority of patients have no known genetic cause, and the underlying molecular mechanisms driving each subtype remain poorly understood. This knowledge gap has made it difficult to distinguish between subtypes in living patients, hampering efforts to predict outcomes or develop targeted treatments.
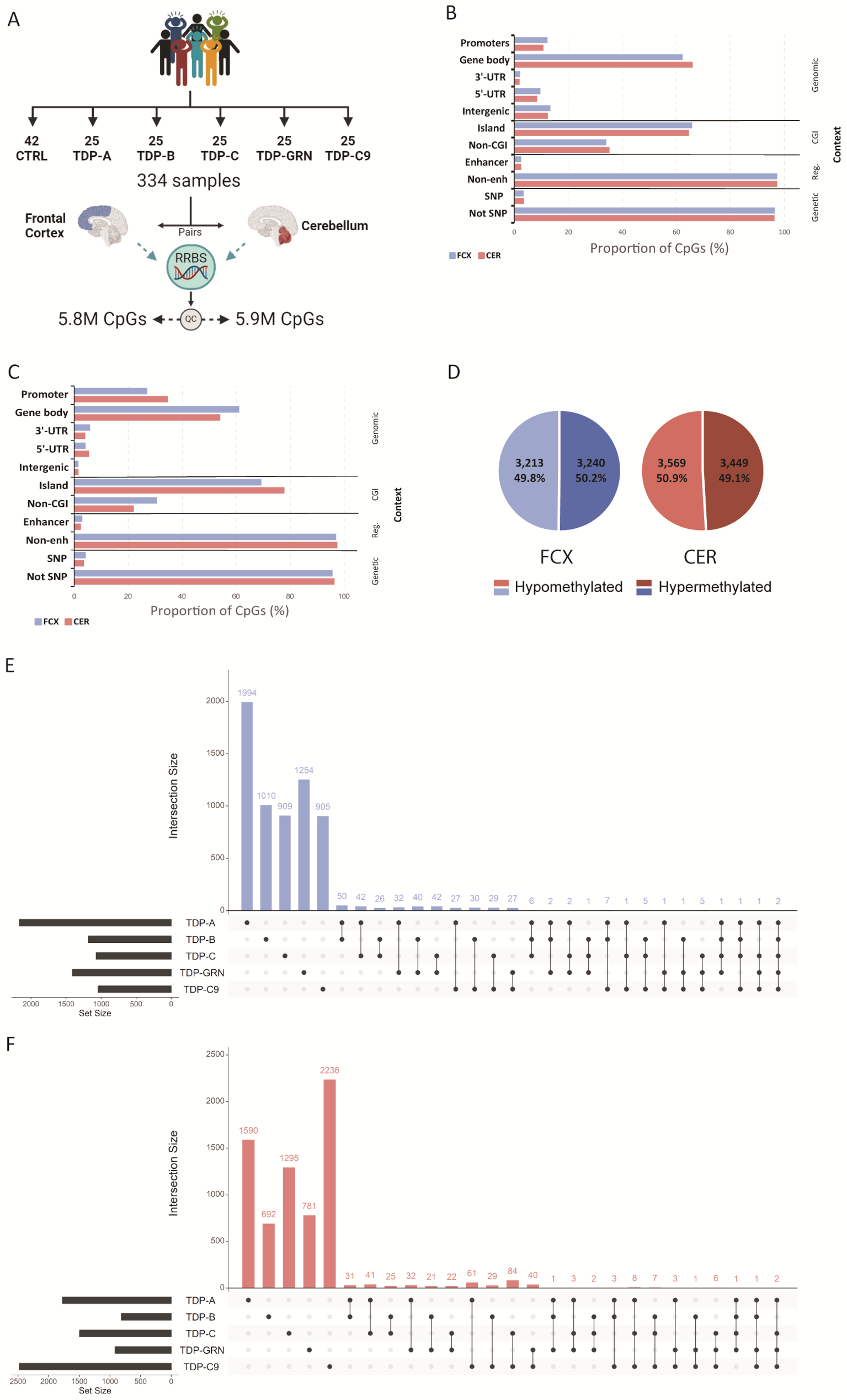
To address this gap, a new study, led by Cristina Vicente investigated epigenetic changes as a promising way to find new risk factors or biomarkers for the disease. The researchers analyzed brain tissue from individuals with genetic forms of the disease as well as sporadic cases using an advanced DNA methylation analysis technique called reduced representation bisulfite sequencing (RRBS).
The team discovered thousands of differentially methylated sites in both the frontal cortex and cerebellum, with over 90% of these changes being unique to specific FTLD-TDP subtypes—TDP-A, TDP-B, TDP-C, GRN mutation carriers, and C9orf72 repeat expansion carriers.
"What's particularly striking is how distinct each subtype appears at the epigenetic level," explains Vicente. “These subtype-specific methylation patterns add another dimension to the molecular signatures that distinguish these conditions, revealing that in each subtype different biological processes are affected in the brain.”
Further analysis of biological processes disrupted by these methylation changes revealed remarkably distinct disease mechanisms. TDP-A patients showed changes in genes involved in protein phosphorylation and DNA repair processes, TDP-B patients had alterations in cholesterol biosynthesis pathways, while TDP-C patients showed disruption in genes controlling protein localization and transcription regulation. These findings suggest that each subtype affects the brain through fundamentally different molecular pathways.
Beyond these broader patterns, the study identified several specific genes with significant methylation changes that could serve as potential biomarkers. Most notably, CAMTA1 showed consistent hypomethylation in TDP-A patients. This methylation change is correlated with reduced CAMTA1 expression and appeared to influence neighboring genes involved in cellular function. The findings suggest a "double-hit" mechanism for CAMTA1 dysfunction in TDP-A patients, where both TDP-43 pathology and epigenetic changes may contribute to disease progression.
The researchers also discovered significant methylation changes in GFPT2, particularly strong in TDP-C patients. This gene plays a crucial role in the hexosamine biosynthesis pathway, which has been linked to preventing harmful protein aggregation—a known hallmark of FTLD-TDP.
"Our findings show that each subtype has completely distinct molecular mechanisms—they're so different they could almost be considered separate diseases entirely."
The implications for patients and families affected by FTLD could be significant. Currently, distinguishing between FTLD-TDP subtypes requires post-mortem brain examination, but these newly identified methylation signatures could potentially be developed into diagnostic biomarkers accessible through blood or other tissue samples during a patient's lifetime.
Furthermore, unlike genetic mutations, epigenetic changes can potentially be reversed with appropriate treatments, offering hope for future beneficial interventions.
"Our findings show that each subtype has completely distinct molecular mechanisms—they're so different they could almost be considered separate diseases entirely," says Rosa Rademakers. "We need to dig deeper into other epigenetic layers to fully understand how each subtype develops and progresses, which will ultimately give us the precision tools needed for better diagnosis and targeted therapies."
Congratulations to Cristina Vicente, the whole team at VIB-UAntwerp Center for Molecular Neurology and all collaborators.