JAXLEY enables large-scale, differentiable simulations of brain activity
Researchers have developed a powerful new software toolbox that brings the tools of modern machine learning to realistic models of brain activity.
Researchers have developed a powerful new software toolbox that brings the tools of modern machine learning to realistic models of brain activity. This open-source framework, called JAXLEY, combines the precision of biophysical models with the scalability and flexibility of modern machine learning techniques. The study, published in Nature Methods, could be a major step toward faster and more accurate simulations of brain function.
Understanding how neurons give rise to thought, perception, or memory remains one of the biggest challenges in neuroscience. To explore these questions, scientists build computer models that simulate the activity of real brain cells. Such biophysical models are digital reconstructions of neuronal networks that include realistic features such as dendrites, synapses, and ion channels. They enable researchers to study how messages travel as electrical signals through individual neurons or across entire networks.
But getting these models to act like real neurons is a technical challenge: each model includes thousands of equations that describe how electrical signals and chemicals move inside brain cells. Adjusting all those parameters to accurately match experimental data is both complex and computationally expensive, often requiring weeks of manual tuning or inefficient trial-and-error simulations.
Differentiable simulations for neuroscience
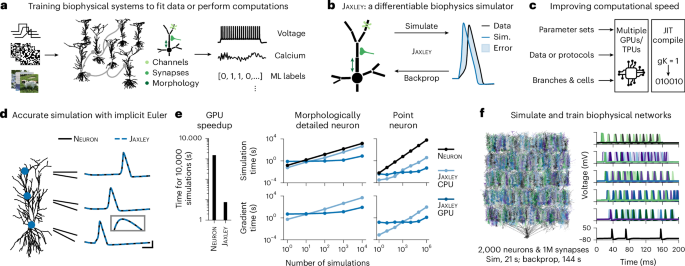
To overcome these limitations, researchers at NERF, VIB.AI, and the University of Tübingen have developed JAXLEY: a new open-source toolbox for differentiable simulations of biophysical neuron and network models. Built on the JAX machine learning framework, JAXLEY allows neuroscientists to compute gradients efficiently and run large-scale simulations on GPUs, unlocking a new level of speed and flexibility.
To demonstrate its versatility, the researchers put JAXLEY to the test on a wide range of tasks. It accurately reproduced experimental voltage and calcium recordings, fitted detailed models with thousands of parameters, and trained large biophysical networks with realistic neuronal and synaptic dynamics. These results show that even complex realistic neuronal networks can now be trained directly on experimental data or computational tasks, something that was previously difficult to achieve.

Pedro Gonçalves (NERF, VIB.AI) co-authored the new paper together with close collaborators at the University of Tübingen.
Bridging neuroscience and machine learning
JAXLEY bridges the gap between traditional neuroscience modelling and modern machine learning. By making biophysical simulations differentiable and GPU-accelerated, it can process large-scale datasets in parallel, increasing the speed and efficiency of brain modelling.
By combining the biophysical precision of mechanistic models with the scalability of AI methods, JAXLEY overcomes computational barriers that have so far limited the scale of brain simulations. The toolbox is fully open-source, making it accessible to the entire research community. This work opens new directions for exploring how the brain learns and remembers, replacing manual tuning with adaptive, data-driven learning inspired by the brain itself.
“JAXLEY fundamentally changes how we approach brain modeling,” says Pedro Gonçalves, group leader at NERF and VIB.AI. “It allows us to build realistic models that can be optimized and scaled efficiently, opening new directions for understanding how neural computations arise from biophysical mechanisms.”