Bring tissue maps to life with Visium HD
At VIB Technologies, Visium HD is part of an end-to-end collaborative pipeline, from tissue processing to spatial data analysis
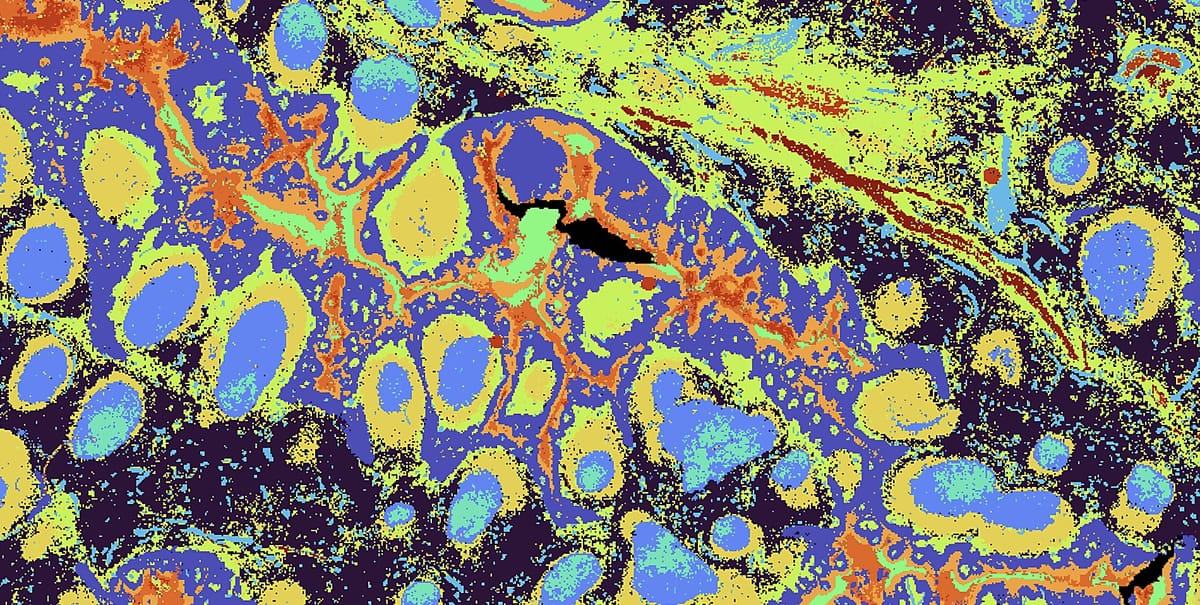
Spatial biology is reshaping how scientists explore tissues, development, and disease. With the launch of Visium HD services through VIB Technologies, researchers have access to untargeted spatial transcriptomics with near single cell resolution, backed by a reliable, integrated workflow developed through close collaboration across multiple VIB Technology platforms.
Booming business
Spatial transcriptomics has grown from coarse tissue maps to near-single-cell resolution, and Visium HD marks a major step forward in this evolution. By capturing RNA directly from tissue sections and mapping expression back onto histology, Visium HD enables researchers to explore tissue architecture at unprecedented detail.
Compared to earlier 50-µm grids, HD pushes to ~2-µm features with higher sensitivity, making it practical to resolve cellular neighborhoods and fine structures. We support both probe-based chemistries and direct-capture options on fresh-frozen material. The result: rich spatial maps matched to your tissue and question, and designed to integrate with single-cell data from the same donor or block.
At VIB, Visium HD has quickly become a high-demand service, with over 100 projects already underway across different research areas, says Evelien Van Hamme, who leads VIB’s Spatial Catalyst:
"Visium HD offers researchers an unprecedented window into tissue architecture. Working together across platforms allowed us to build a solution that’s technically sound, scalable, and already driving discovery."
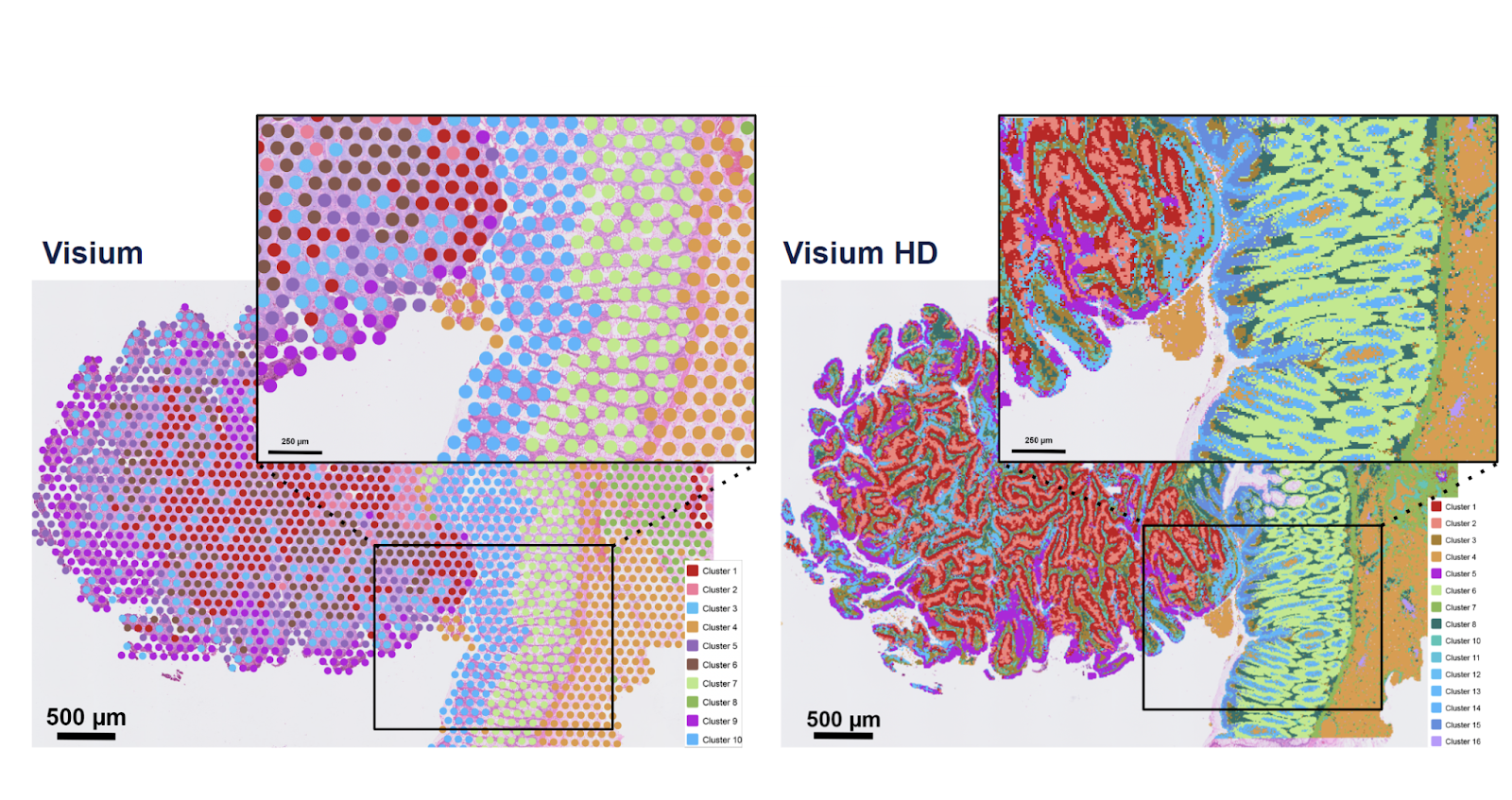
In a nutshell, users get:
- Spatially resolved expression tied to anatomy and pathology, not just clusters
- A project co-designed for single-cell/single-nucleus integration when useful
- End-to-end traceability (every sample, slide, run, and QC is recorded)
- Cost-efficient, depth-matched sequencing runs
- Standardized outputs plus optional expert analyses
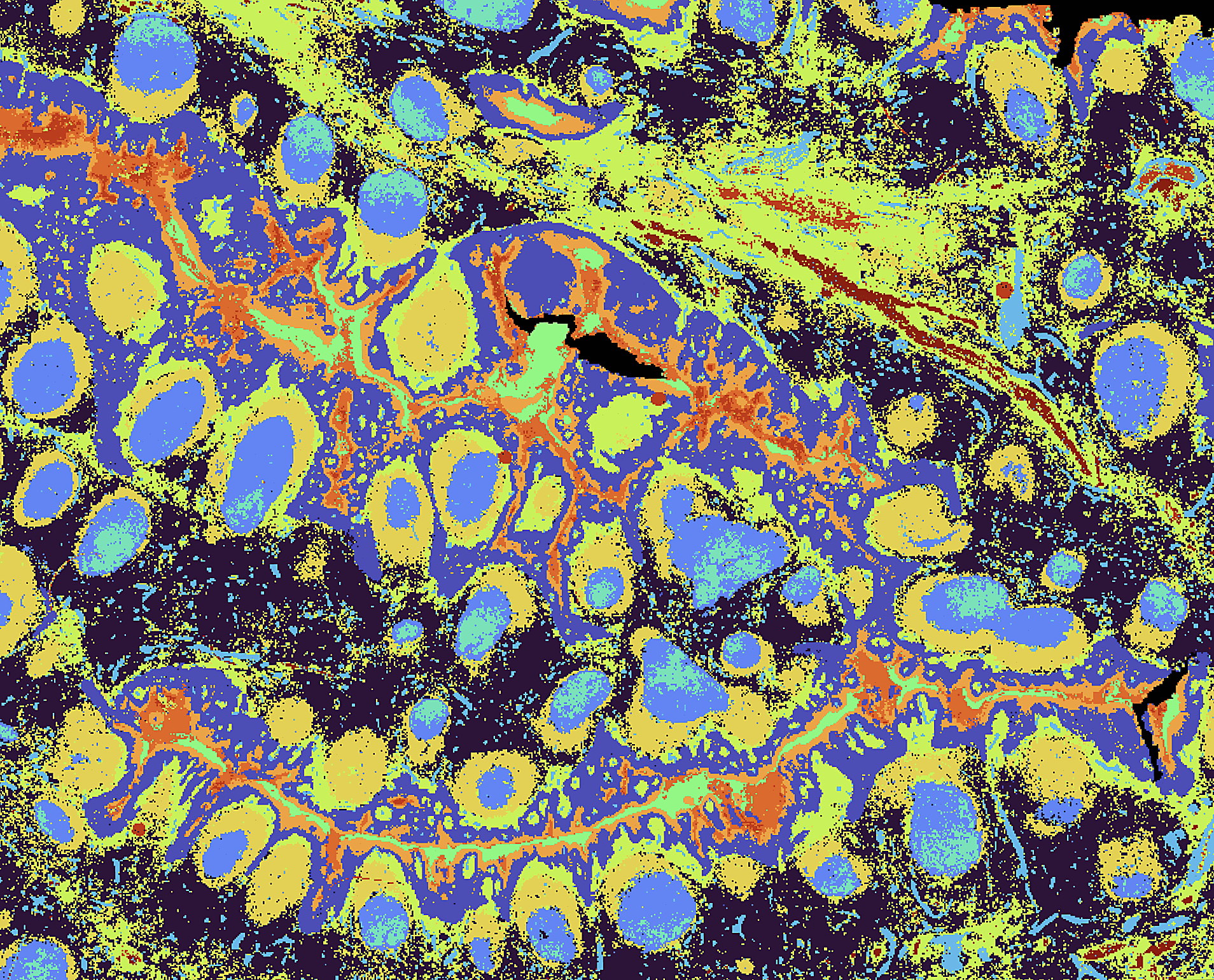
One streamlined pipeline
- Project design
Together we decide: fresh-frozen vs FFPE; probe-based vs direct capture; whether to add sc/snRNA-seq from the same donor or adjacent sections; and what imaging/coverage you’ll need. Getting this right upfront enables clean downstream integration. - Tissue intake & QC
We handle embedding, sectioning, storage, and RNA QC to confirm tissue suitability before irreversible steps. We optimize sectioning and mounting for spatial capture, and align parallel single-cell work if planned. - Fast histology imaging
We image the sections promptly, creating the anatomical scaffold your expression maps will register to. - Spatial capture (CytAssist + Visium HD slide)
Probe hybridization or direct capture happens on barcoded Visium HD slides using the CytAssist device. This is the critical “location-encoding” step. SOPs are tightly standardized so every pixel’s barcode faithfully maps to tissue. - Library preparation hand-off
Immediately after capture, samples go for library construction. Hand-off is frictionless because all metadata travel via our shared portal and QC thresholds and acceptance criteria are harmonized across teams. - Sequencing
Libraries are pooled strategically to achieve target depth efficiently and sequenced on high-throughput platforms. Run performance metrics and raw data are automatically linked back to each spatial experiment. - Primary processing & delivery
Pipeline processing starts as soon as data land. You receive tissue-aligned expression matrices, QC reports, and browseable outputs. All slide/kit/run details are recorded so your Materials & Methods section practically writes itself.

Custom analysis
While the Visium HD pipeline delivers high-quality spatial transcriptomics outputs, many projects do not stop there. Once researchers start exploring the data, new questions often emerge that go beyond default deliverables. Addressing those questions requires deeper, custom analysis, and this is where an additional, dedicated service layer comes into play.
On top of the core pipeline, we offer advanced, custom spatial data analysis that allows you to re-engage with your datasets in a more exploratory and hypothesis-driven way. This second layer builds directly on the standardized outputs but is tailored to the biological challenge at hand.
Key challenges addressed at this stage include:
- Cell and nucleus segmentation in complex tissues
- Accurate co-registration between histology, spatial transcriptomics, and other imaging modalities
- Resolving cellular neighborhoods and spatial gradients beyond spot-level summaries
- Integration with matched single-cell or single-nucleus datasets
- Region-of-interest–based comparisons and spatially informed differential expression
- Inference of cell-cell interactions and signaling niches
Significant effort has gone into developing spatial analysis software from a data-first perspective, tightly coupled to real experimental workflows. Tools such as Harpy and Sparrow have been developed and iteratively refined alongside wet-lab protocols, ensuring that computational methods keep pace with the resolution and complexity of Visium HD data.
This intertwined development of wet-lab workflows and bioinformatics infrastructure is a distinguishing feature of the VIB spatial ecosystem.
Geert Van Minnebruggen: “While many institutes have established spatial transcriptomics protocols at the bench, far fewer have invested at the same depth in robust, scalable, and reusable data analysis frameworks designed specifically for high-resolution spatial data.”



Evelien Van Hamme, Isabelle Scheyltjens and Niels Vandamme
Enabled by teamwork
In practice, this means Visium HD at VIB is not a single pipeline, but a two-tier offering: a reliable service for spatial data generation, followed by a flexible, custom analysis layer that turns spatial datasets into biological understanding.
None of this would be possible without the joint effort of different VIB Cores, each bringing unique strengths to the table.
“The Spatial Catalyst took the lead in benchmarking the technology and brings deep expertise in tissue preparation, imaging workflows, and spatial platforms including Xenium, NovaST, BMKGene, MACSima, and more,” says Isabelle Scheyltjens. “Their bioinformatics team is instrumental in software development and troubleshooting.”
In parallel, the Single Cell Core contributes its strengths in molecular biology, sequencing strategy, and workflow design, drawing from years of experience in high-throughput projects and multi-omics assays.
“The Single Cell Core also built data workflows for pre-processing and data management, bridging wet lab and computational steps ensuring users receive data that is not only high-resolution, but also analysis-ready.”
Nucleomics Core contributes its high-throughput sequencing expertise and infrastructure, and the BioImaging Cores support swift imaging.
But beyond the technology, what truly defines this pipeline is the way it was built: not by individual units working in isolation, but through collaborative platform integration, a model that VIB Technologies is actively developing and scaling.
Niels Vandamme, Head of the VIB Single Cell Core: "This is what the future looks like: not cores working in isolation, but expert teams collaborating across boundaries. Together, we created something greater than the sum of its parts, and researchers benefit from that unified approach."
Contact Isabelle Scheyltjens, Visium HD project manager
You can also reach the involved platforms via the VIB Technologies website: connect.vib.be
